Third
International Electronic Conference on Synthetic Organic Chemistry (ECSOC-3),
www.reprints.net/ecsoc-3.htm, September 1-30, 1999
[E0004]
JChemPaint
- Using the Collaborative Forces of the Internet to Develop a Free Editor
for 2D Chemical Structures.
MPI of Chemical Ecology, Tatzendpromenade 1a, 07745 Jena, Germany
E-mail: [email protected]
* Author to whom correspondence should be addressed
Received: 17 August 1997 / Uploaded: 21 August 1997
Abstract
The open source program JChemPaint for drawing 2D chemical structure, its
current features, its envisioned further development and the principles
enabling researchers and students at places all over the world to collaboratively
develop such a program are described.
Introduction
2D chemical structure editors are central tools for chemoinformatics and
computational chemistry. No matter if one wants to submit a structure query
to a database, prepare a starting structure for molecular modeling, draw
a set of structures for goodlists and badlists in Computer Assisted Structure
Elucidation (CASE) or just sketch a reaction scheme for a publication -
in any of these cases the starting point is opening a structure editor.
Programs for drawing chemical structures are abundant and a number of formerly
commercial programs in this area are now available free of charge for non-profit
use, like
Isis Draw. Nevertheless, there
are no state-of-the-art programs available in source code with a free licensing
scheme, which enable researchers to adapt and embed them into their own
programs without paying license fees. Such an open source structure editor
would be of interest for many reasons. Just not having to pay for
it is certainly the weakest argument. Firstly, it would ease the work of
all those developers who need to be able to change and adapt the source
code of a module they use in order to integrate it into their projects.
The makers of a programs that calculate NMR shifts for a given structure
or generate the IUPAC name would not have to rewrite this standard piece
of software again and again. Secondly, bugs are much more easily found
and improvements are much more easily made if everyone can have a look
at the source code.
Thus, the intriguing characteristics of the open
source paradigm, the introduction of Java
with its platform independence as well as the surprising lack of a free,
open source, platform independent structure editor made it desirable to
start the JChemPaint project.
Program description
JChemPaint is a program for drawing 2D chemical structures, written in
Java. We decided to use Java because of its unique features of being platform
independent, easy to learn, highly structured and well integrated with
web technology, enabling the use of JChemPaint for all kinds of web based
projects.
JChemPaint (Figure 1), currently supports:
-
A subset of the regular drawing features of commercial programs, as there
are
-
drawing of single, double and triple bonds (no stereo descriptors yet).
-
deletion of bonds and atoms
-
ring templates (3-8)
-
one click attachment of ring templates to an atom or a bond
-
flipping and rotating selected parts of the molecule
-
Loading and saving of structures as MDL Molfiles and in Chemical Markup
Language (CML).
-
Automated Structure Layout, also known as Structure Diagram Generation.
Taking into account the fair number of programs available in this field,
there seem to be no greater challenges in designing such a system. However,
some aspects are nice and tricky and pose interesting problems e.g. for
student education. An object oriented system like JChemPaint, with its
clear and modular design, and its source code available to everyone, seems
to be the ideal playground for trying new ideas and optimizing existing
solutions.
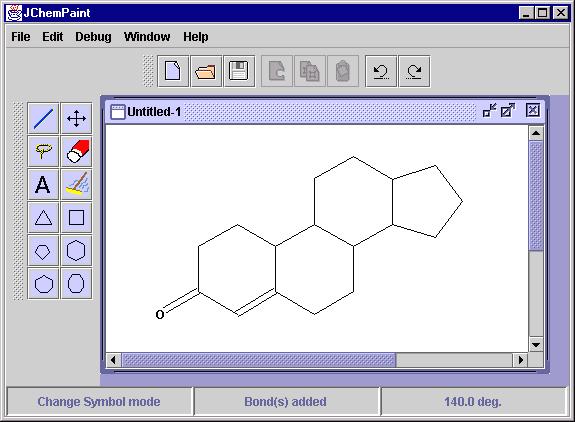
Figure 1: A screen shot of JChemPaint
The most prominent solution for which an implementation in JChemPaint
exists is the one for the problem of Structure Diagram Generation - comprehensively
summarized in the review of Harold E. Helson at CambridgeSoft [1].
Here, a molecular graph, either without any layout information or with
some layout characteristics that make a cleanup desirable, is subjected
to an algorithm which places each atom in the molecule such that the resulting
picture of the molecule complies with the conventions used by chemists
to hand draw such structures. In JChemPaint we use a the Java module JMDraw
written by one of us (CS) which is based on the C program MDraw by Ugi
and coworkers [2]. While the resulting layout is sufficient
in many cases there is still plenty of room for improvements and JChemPaint's
open source is the ideal basis for that.
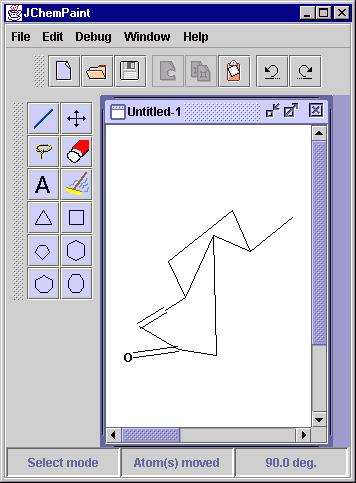
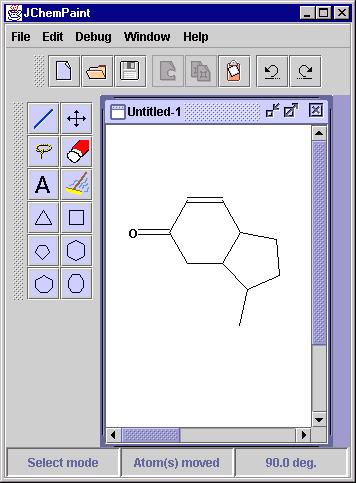
Figure 2: Before and after - the effect of a JMDraw clean-up.
Another case for using JChemPaint for educational purposes is its capability
of handling Chemical Markup Language (CML), the upcoming universal language
for managing chemical information [3]. CML is an extension
of XML, the Extensible
Markup Language, and is likely to have a large impact on the
the way of how chemists encode their chemical information. The process
of designing CML is not yet closed and it is thus especially existing to
have look at or even take part in the ongoing development. For more details
on CML and its implementation in JChemPaint and JMol please see Egon
Willighagens article on "Processing CML conventions in Java" and the
references therein.
The Development Model
The JChemPaint project was started by Christoph
Steinbeck and Stefan Krause
from the ChemoInformatics
group at the Max Planck
Institute of Chemical Ecology in Jena. It was soon discovered that
a complementary 3D program, JMol,
was developed within the Open Science
Project of Dan Gezelter
at Columbia University. It quickly became our vision that both programs
should form a comprehensive system for 2D and 3D handling of chemical structures
like found for example in the commercial ChemOffice suite. Egon
Willighagen has joined both teams and added support for structure I/O
in CML (Chemical Markup Language). New versions of the program are released
frequently and early, as recommend by Eric Raymond in his brilliant analyses
of the principles driving the open source development. Each announcements
causes a number of new interested potential co-developers to join the developers
mailing lists and a number of them contributes by discussing questions
of program design. The development of JChemPaint is maintained via the
Concurrent Versions System (CVS) system,
a widely used computer program with a client-server architecture that allows
users to independently and concurrently work on even the same parts of
the source code by checking out personal copies of the software from the
central repository, making their changes and checking in again the modified
source code. The CVS system then tries to merge the independently modified
versions of the source into the repository and does only in rare cases
require intervention by the user for this purpose. Communication between
the developers is organized via the JChemPaint web pages and electronic
mailing lists, one for the program's users and one for its developers.
Conclusion and Outlook
We have described the program JChemPaint, a 2D molecular structure editor.
While the program itself as well as most of the underlying algorithms are
no scientifically thrilling material, it is its development model and the
wide usability of the program that might attract the attention of a potentially
large group of users and of some highly welcome new co-developers.
A great number of improvements and new features waits for implementation.
Professional quality outputs are not possible at this time nor is adaptation
to different types of layouts, to mention only two possible fields of potential
development. A lot of work is also to be done in the area of interfacing
the program with JMol. Here, a 3D model builder, the 3D analogue of JMDraw,
is the first thing to mention, which, needless to say, is a whole new open
source project by itself. The experiences from other open source
developments show that a critical mass of working features must implemented
in order to attract contributors. We hope that, as the program grows, the
community also will.
References
[1] H. E. Helson, in Reviews in Computational Chemistry, K. B. Lipkowitz
and D. B. Boyd, Eds., Wiley-VCH, New York, 1999, Vol. 13, pp. 313-398.
Structure Diagram Generation.
[2] K. Bley, J. Brandt, A. Dengler, R. Frank and
I. Ugi, "Constitutional Formulae generated from
Connectiviy Information: the Program MDRAW", Journal of Chemical Research
(M), 1991, 2601-2689.
[3] For documentation of Chemical Markup Language
(CML) please see www.xml-cml.org.
All comments on this poster should be sent by e-mail to (mailto:[email protected]
ona.edu) [email protected]
with E0004 as the message subject of your e-mail.