

[C0026]
A New Receptor Molecule for Lysine and Histidine in Water – Strong Binding of Basic Amino Acids by a Macrocyclic Host
Thomas Grawe,a Thomas Schrader*,b Paolo Finocchiaro*,c Salvatore Failla,cGiuseppe Consigliod
aInstitut für Organische Chemie und Makromolekulare Chemie, Heinrich-Heine-Universität Düsseldorf, Universitätsstr. 1, D-40225 Düsseldorf, Germany
bFachbereich Chemie, Philipps-Universität Marburg, Hans-Meerwein-Straße, D-35032 Marburg, Germany
cDipartimento di Ingegneria Chimica, dei Materiali delle Materie Prime e
Metallurgiche-Università di Roma la Sapienza-Via del Castro Laurenziano,7-00161 Roma, Italy
dUniversitá di Catania, Facoltá di Ingegneria, Istituto Chimico,
Cittá Universitaria, Viale A. Doria, I-95125 Catania, Italy
E-mail: [email protected]
Received: 7 August 2000 / Uploaded: 11 August
Introduction
The effective and selective molecular recognition of amino acids in water is still a challenge in supramolecular chemistry. Because of the frequent use of the side chains of basic amino acids (Lys, Arg, His) for biological processes the molecular recognition of these amino acids by synthetic receptor molecules is of special interest [1]. Recently we presented a new macrocyclic receptor molecule, which binds arginine and lysine in a stereoselective fashion (Fig. 1) [2]. The mechanism of enantioselective recognition relies on two simultaneous cation-phosphonate interactions – i. e. binding of a specific cationic group of the amino acid to a specific phosphonate moiety in the receptor molecule. After docking the amino acid comes into van der Waals-contact of the chiral surface of the chiral bridging unit in 1 and one enantiomer is bound preferentially.
|
|
Fig. 1. Chiral recognition of lysine by macrocyclic bisphosphonate receptor molecule 1 in DMSO.1
However, because of the relatively weak binding energy from the classical ammonium phosphonate interaction the overall binding constants were in the range of 104 M-1 only in DMSO. We asked ourselves, if strong binding in more polar solvents (preferably in water) could be achieved, if instead of two single phosphonates two chelating bisphosphonate moieties would be incorporated in a macrocycle, so that each cationic functionalitiy of the amino acid would be involved in a highly effective chelate complex. This lead to the design of macrocycle 2 which according to X-ray analysis and molecular modeling adopts a favorable open conformation [4]. The respective tetraanion should be able to perform an induced fit on approach of a dication with the correct spacer. Thus, a highly stable 1:1-complex could be formed, possibly even in water (Fig. 2).
 |
|
Fig. 2. Tetrakisphosphonate rceptor molecule 3 and its complex with lysine methylester dication (proposed binding mode). Left: Lewis structures; right: molecular mechanics calculations (cylinders: arginine, CPK: host 3).
Discussion
The synthesis of macrocycle 2 and some related structures has recently been described by the Finocchiaro-group [3].3 In a very efficient one-pot synthesis 2,6-bis(bromomethyl)pyridine is directly treated with phosphonate-modified bisphenol A in a dipolar aprotic solvent with a mild base. Macrocycle 2 is obtained in 70% yield by simultaneous formation of four C-O-bonds. For mild and selective monodealkylation of all four dimethylphosphonates we modified a procedure originally described by Karaman et al [5]. Heating of an acetonitrile solution of 2 with 4 equivalents of dry lithium bromide for 1 week afforded, after recrystallization and dialysis, the pure receptor molecule 3 as the tetralithium salt. This compound is well soluble in polar solvents such as methanol and water, but insoluble in DMSO and acetonitrile.

Fig. 3. Synthesis of macrocyclic tetraphosphonate receptor molecule 3.
In a preliminary experiment, a 1:1-mixture of 3 and lysine methylester dihydrochloride produced significant complexation induced shifts of CH-protons both in the amino acid as well as in the receptor molecule. To check the complex stoichiometries with lysine, arginine, orhithine and histidine, Job-Plots were taken for each of the amino acid-complexes with 3 [6]. The result was remarkable: While the smaller amino acids histidine, ornithine and arginine produced a clear 2:1-stoichiometry, only lysine was bound by 3 in a clean 1:1-complex.
 |
 |
Fig. 4. Job-plots for the complex between 3 and histidine (left) or lysine methyl ester (right); for the proton numbering scheme of 3 see Supporting Information.
This is a good indication for the postulated binding mode: Only in lysine the two cationic groups are able to span the distance from one bisphosphonate moiety to the other [2].2 By contrast, the smaller amino acids arginine, ornithine and histidine can only reach one bisphophonate moiety at a time, so that two amino acid molecules can be bound by receptor molecule 3. However, even in this case a relatively stable assembly may be formed, characterized by an array of alternating positive and negative charges according to energy minimizations [7]. Histidine is bound remarkably well in this 2:1-complex. The reason for this may lie in the special hydrogen bond donor-arrangement of the imidazolium ion: According to force-field calculations two histidine molecules can each be involved in a double chelate assembly with three phosphonate groups. While the ammonium functionality is complexed as usual by the bisphosphonate, the imidazolium ring acts as a hydrogen bond donor-bridge to one of the phosphonate moieties at the opposite end of the receptor molecule (Fig. 5) [8].
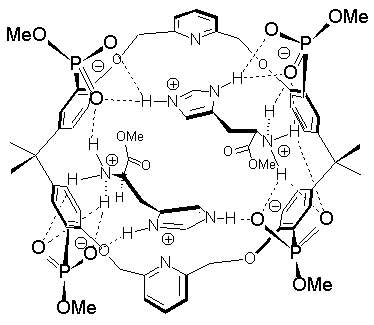 |
 |
Fig. 5. Postulated binding mode of histidine methyl ester in its 2:1-complex with tetraphosphonate 3 (energy-minimization).
We then performed NMR titrations for each of the four complexes in methanol. The resulting binding curves were analyzed by nonlinear regression; the respective association constants are listed in table 1 [9].
 |
   |
 |
   |
Fig. 6. NMR titration curves for the complexation of histidine (left) or lysine methyl ester (right) by 3.
In methanol all amino acids are bound strongly by 3. The four-point interaction in lysine’s complex with 3 is more powerful than the two-point interaction in the related assemblies with ornithine and arginine. However, in spite of its 2:1-stoichiometry, histidine is even superior to lysine. This is again in good agreement with the above discussed double chelate binding mode suggested by molecular modelling. In addition to the enhanced electrostatic attraction, histidine is able to form two strong hydrogen bonds between the imidazolium and and the bisphosphonate ions. This may contribute to its superior binding constant.
From methanol to water, the stoichiometry of all complexes is retained, but a 20-50 fold drop is observed in the association constants of the four investigated amino acids. This demonstrates the powerful competition of water molecules and is in accord with the experience made by other groups. However, this time, lysine is complexed 5-7 times more strongly than ornithine and arginine and even twice as strongly as histidine. If hydrophobic forces are weak as in our case, the contribution of hydrogen bonds in water is negligible, while electrostatic interactions represent the major attractive force. It is known, that in this respect the hard ammonium ion with its high charge density is superior to the softer guanidinium and also the imidazolium ion, where the positive charge is delocalized across several atoms [10]. In our case, the electrostatic attraction exerted by the second ammonium functionality of lysine is obviously much stronger than that of arginine’s guanidinium ion and even histidine’s imidazolium ion. Thus, receptor molecule 3 is moderateley selective for lysine in water.

Figure 7. Investigated dicationic guest-molecules: the distance between the charged nitrogen atoms is successively increased by one carbon or nitrogen atom.
Table 1. Association constants (Ka)[M-1] and stoichiometries for the complexes of basic amino acids with receptor molecule 3 from NMR titrations in methanol and water at 20°C.[a]
|
Amino acid dihydrochloride |
(Ka) [M-1](methanol) |
Stoichiometry[b] (Amino acid:3) |
(Ka) [M-1](water) |
Stoichiometry[b] (Amino acid:3) |
|
|
||||
|
Histidine |
29000 ± 11% |
2:1 |
650 ± 18% |
2:1 |
|
Ornithine |
9500 ± 9% |
2:1 |
221 ± 33% |
2:1 |
|
Arginine |
8800 ± 14% |
2:1 |
165 ± 38% |
2:1 |
|
Lysine |
21000 ± 35% |
1:1 |
1200 ± 25% |
1:1 |
a
Due to the strongly hygroscopic character of both titration partners the d4-methanol solution contained ~0.1% of water. Errors in Ka are standard deviations and were calculated to be ± 15-38%; b from Job-Plots.Recently, Bell [11] and Dougherty [12] have published new binding motifs for arginine, which both operate very efficiently in water. While Bell’s rigid halfmoon-shaped receptor molecule is highly preorganized, Dougherty makes use of the p-cation interaction between electron-rich benzene rings and the guanidinium moiety, enforced by multiple electrostatic interactions with additional carboxylates in the periphery of the receptor molecule. However, both hosts are large molecules, accessible only through a multistep synthesis. In this respect our modular synthesis from two building blocks in one step may offer the advantage of a fast entry into a whole family of related tetraphosphonate hosts.
Conclusions and Outlook
In the future, we intend to incorporate chiral building blocks in our receptor molecules (preferably fom natural sources such as amino acids) as well as implement more rigid elements for a high degree of preorientation to achieve enantioselective and strong recognition of basic amino acids in water.
Experimental Section:
General Methods: Deuterium oxide (Merck) and methanol-d4 (Merck) were purchased each in 99.8% purity. 2-hexanone (Aldrich) and lithiumbromide (Merk) were also purchased each in 99% purity.
17,17,40,40-Tetramethyl-1,10,24,33-tetraoxy[2](2,6)pyridino[2.1]paracyclo[2](2,6)-pyridino[2.1]paracyclophane- 12,20,35,43-tetraphosphonic acid monoethylester tetra lithium salt. 146.7 mg (0.1215 mmol) of the macrocycle, 46.3 mg (4.1 eq.) of lithiumbromide were dissolved in 7 ml of dry 2-hexanone under argon atmosphere and heated for 120 hours at 100°C. The residue was filtrated and washed with 2 hexanone. The salt was purified via dialysis; afterwards it contained only minute amounts of lithiumbromide. Yield: 116 mg (85%). 1H-NMR (500 MHz, CD3OD): d = 1.13 (t, 12 H, J = 7.3 Hz, CH3CH2O), 1.64 (s, 12 H, CH3C), 3.81 (q, 8 H, J = 7.0 Hz, CH3CH2O), 5.26 (s, 8 H, CH2O), 6.76 und 6.90 (m, 2 x 4 Harom.), 7.69 (d, 4 Hpy, J = 6.9 Hz), 7.76 (t, 2 Hpy, J = 6.9 Hz), 7.86 (m, 4 Harom.). 13C-NMR (125 MHz, CD3OD) d = 17.44 (s), 31.87 (s), 39.00 (s), 43.37 (s), 61.74 (d, 5.0 Hz), 113.75 (d, 8.8 Hz), 122.36 (s), 123.92 (s), 125.31 (s), 129.36 (d, 82.3 Hz), 132.39 (s), 133.58 (d, 6.8 Hz), 139.68 (s), 144.36 (d, 12.5 Hz), 159.01 (d, 42,3 Hz). 31P-NMR (200 MHz, CD3OD) d = 12.85. Anal. Cacld for C52H58N2O16P4Li4: C 55.83; H 5.23; N 2.50. Found: C 54.71; H 5.45; N 2.08. MS (FAB, glycine matrix, Xe): m/z 1125 (M + Li+, 55%).
1
H NMR Titrations.Ten NMR tubes were filled each with 0.8 mL of a solution of the host compound (cguest = 0.5–4 mM) in a deuterated solvent (methanol-d4, or D2O). The guest compound (1.525 equiv corresponding to the guest) was dissolved in 0.61 mL of the same solvent, and the resulting solution was added with increasing volumes from 0 to 5 equiv. to the guest solution in ten NMR tubes.
Volume and concentration changes were taken into account during analysis. The association constants were calculated by non-linear regression methods.
Job plots. Equimolar solutions (10 mmol/10 mL, approx. 10 m M) of dication and tetraphosphonate were prepared and mixed in various amounts. 1H NMR spectra of the mixtures were recorded, and the chemical shifts were analysed by Job’s method modified for NMR results.
Supporting Information:

1. NMR titrations in methanol
 |
   |
 |
  |
 |
   |
 |
   |
2.Job-Plots in methanol
 |
  |
 |
4
|
 |
4
|
 |
4
|
3. NMR titrations in water
 |
1
|
 |
1
|
 |
1
|
 |
1
|
4. Tables
Table 1: Histidine in methanol
|
Eq Guest |
Dd H1 |
Dd H2 |
Dd CH3 |
Ka /Error (averaged) |
|
|
0,25 |
0,2093 |
0,0709 |
0,0255 |
||
|
0,5 |
0,3518 |
0,1235 |
0,0472 |
||
|
0,75 |
0,3934 |
0,1421 |
0,0567 |
||
|
1 |
0,4047 |
0,1475 |
0,0595 |
||
|
1,25 |
0,4084 |
0,1496 |
0,061 |
||
|
1,5 |
0,4097 |
0,1518 |
0,0614 |
||
|
2 |
0,4122 |
0,1516 |
0,0624 |
||
|
3 |
0,4132 |
0,155 |
0,0626 |
Ka =29000 |
|
|
5 |
0,4141 |
0,1551 |
0,0629 |
Error:+/-11% |
Table 2: Ornithine in methanol:
|
Eq Guest |
Dd H1 |
Dd H2 |
Ka /Error (averaged) |
|
|
0,25 |
0,0799 |
0,0351 |
||
|
0,5 |
0,1242 |
0,0592 |
||
|
0,75 |
0,1439 |
0,0715 |
||
|
1 |
0,1533 |
0,0783 |
||
|
1,25 |
0,1569 |
0,0821 |
||
|
1,5 |
0,1596 |
0,0846 |
||
|
2 |
0,1639 |
0,0875 |
||
|
3 |
0,1647 |
0,0895 |
Ka =18000 |
|
|
5 |
0,1673 |
0,0908 |
Error:+/-30% |
Table 3: Arginine in methanol
|
Eq Guest |
Dd H1 |
Dd H2 |
Dd H3 |
Ka /Error (averaged) |
|
|
0,25 |
0,02606 |
0,0347 |
0,04076 |
||
|
0,5 |
0,0405 |
0,0558 |
0,065 |
||
|
0,75 |
0,05018 |
0,0704 |
0,0905 |
||
|
1 |
0,052 |
0,0751 |
0,1018 |
||
|
1,25 |
0,0533 |
0,0804 |
0,1065 |
||
|
1,5 |
0,0541 |
0,0832 |
0,1094 |
||
|
2 |
0,0583 |
0,0874 |
0,1103 |
||
|
3 |
0,06 |
0,0942 |
0,1122 |
Ka = 30000 |
|
|
5 |
0,065 |
0,1012 |
0,1129 |
Error: +/-15% |
Table 4: Lysine in methanol
|
Eq Guest |
Dd H1 |
Dd H2 |
Dd H3 |
Ka /Error (averaged) |
|
|
0,25 |
0,0307 |
0,044 |
0,0632 |
||
|
0,5 |
0,0512 |
0,084 |
0,0999 |
||
|
0,75 |
0,0627 |
0,1005 |
0,1258 |
||
|
1 |
0,0701 |
0,116 |
0,1371 |
||
|
1,25 |
0,0758 |
0,1227 |
0,1455 |
||
|
1,5 |
0,0797 |
0,1298 |
0,1515 |
||
|
2 |
0,0844 |
0,1388 |
0,1609 |
||
|
3 |
0,0896 |
0,1489 |
0,1677 |
Ka = 21000 |
|
|
5 |
0,0934 |
0,1557 |
0,1733 |
Error +/-35% |
Table 5: Histidine in water
|
Eq Guest |
Dd H1 |
Dd H2 |
Dd CH3 |
Dd H4 |
Dd H5 |
Dd H6 |
Dd H2 |
Ka/ Error (averaged) |
|
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
|
|
0,25 |
0,0255 |
0,006 |
0,0063 |
0,0161 |
0,0275 |
0,0081 |
0,0126 |
|
|
0,5 |
0,0637 |
0,0148 |
0,0145 |
0,0432 |
0,0489 |
0,0204 |
0,0312 |
|
|
0,75 |
0,0977 |
0,0249 |
0,0258 |
0,0693 |
0,0489 |
0,0296 |
0,0479 |
|
|
1 |
0,1274 |
0,0384 |
0,0283 |
0,0864 |
0,0773 |
0,039 |
0,0624 |
|
|
1,25 |
0,1649 |
0,0428 |
0,0428 |
0,1166 |
0,0814 |
0,0501 |
0,0801 |
|
|
1,5 |
0,197 |
0,052 |
0,0514 |
0,1359 |
0,0971 |
0,0589 |
0,0946 |
|
|
2 |
0,232 |
0,0595 |
0,0592 |
0,1604 |
0,1126 |
0,0671 |
0,1087 |
|
|
3 |
0,2827 |
0,0718 |
0,0681 |
0,1951 |
0,1349 |
0,0778 |
0,1273 |
Ka = 650 M-1 |
|
5 |
0,3508 |
0,0844 |
0,0835 |
0,2235 |
0,162 |
0,0879 |
0,1456 |
Error = +/- 18% |
Table 6: Ornithine in water
|
Eq Guest |
Dd H1 |
Dd H2 |
Dd CH3 |
Dd H4 |
Dd H5 |
Dd H6 |
Ka/ Error (averaged) |
|
|
0,25 |
0,01 |
0,0019 |
0,0021 |
0,0049 |
0,0055 |
0,0061 |
||
|
0,75 |
0,0231 |
0,0048 |
0,0048 |
0,0108 |
0,0115 |
0,0157 |
||
|
1 |
0,0344 |
0,0075 |
0,0081 |
0,0169 |
0,0169 |
0,0234 |
||
|
1,25 |
0,0414 |
0,0121 |
0,0122 |
0,0235 |
0,0235 |
0,0309 |
||
|
1,5 |
0,0552 |
0,0122 |
0,013 |
0,0275 |
0,0284 |
0,0376 |
||
|
2 |
0,0674 |
0,0165 |
0,0165 |
0,0344 |
0,0343 |
0,047 |
||
|
3 |
0,0848 |
0,0206 |
0,0212 |
0,0432 |
0,0441 |
0,0596 |
||
|
5 |
0,1109 |
0,0272 |
0,0269 |
0,0566 |
0,0566 |
0,0781 |
Ka = 450 |
|
|
6 |
0,12 |
0,03 |
0,0296 |
0,0615 |
0,0611 |
0,083 |
Error = +/- 33% |
Table 7: Arginine in water
|
Eq Gast |
Dd H1 |
Dd H2 |
Dd CH3 |
Dd H4 |
Dd H5 |
Ka/ Error (averaged) |
|
|
0,25 |
0,0074 |
0,004 |
0,0036 |
0,0051 |
0,0037 |
||
|
0,5 |
0,0156 |
0,0084 |
0,0082 |
0,0109 |
0,007 |
||
|
0,75 |
0,0328 |
0,0175 |
0,0164 |
0,0229 |
0,0139 |
||
|
1 |
0,0543 |
0,03 |
0,0317 |
0,0401 |
0,0204 |
||
|
1,25 |
0,0632 |
0,0328 |
0,0317 |
0,0457 |
0,0254 |
||
|
1,5 |
0,0772 |
0,0396 |
0,0395 |
0,0549 |
0,0308 |
||
|
2 |
0,0989 |
0,0508 |
0,0498 |
0,0712 |
0,0383 |
||
|
3 |
0,1316 |
0,0658 |
0,0659 |
0,0939 |
0,0508 |
||
|
5 |
0,1847 |
0,0911 |
0,0911 |
0,1292 |
0,0722 |
Ka = 350 |
|
|
6 |
0,205004 |
0,10054 |
0,101 |
0,145 |
0,077 |
Error: +/-38% |
Table 8: Lysine in water
|
Eq Guest |
Dd H1 |
Dd H2 |
Dd CH3 |
Ka/ Error (averaged) |
|
|
0,25 |
0,0077 |
0,0068 |
8,00000E |
||
|
0,5 |
0,0126 |
0,0104 |
0,0011 |
||
|
0,75 |
0,0157 |
0,0121 |
0,0017 |
||
|
1 |
0,0197 |
0,0155 |
0,0021 |
||
|
1,25 |
0,0231 |
0,0178 |
0,00245 |
||
|
1,5 |
0,0255 |
0,0205 |
0,0028 |
||
|
2 |
0,0315 |
0,0252 |
0,0032 |
||
|
3 |
0,0398 |
0,0314 |
0,004 |
||
|
5 |
0,0532 |
0,0422 |
0,0052 |
||
|
6 |
0,059 |
0,04601 |
0,005705 |
||
|
Ka |
1061,388 |
1135,354 |
1411,877 |
||
|
Dd sat |
0,1052 |
0,080483 |
0,009067 |
Ka = 1200 |
|
|
Error [%] |
+/-23,44 |
+/-26,05 |
+/-25,93 |
Error +/-25% |
5. Job-Plots in water
 |
1
|
 |
1
|
References
[1] The first receptor molecules for arginine derivatives: J.-M. Lehn, P. Vierling, R. C. Hayward, J. Chem. Soc. Chem. Commun.1979, 296-298; A. V. Eliseev, M. I. Nelen, J. Am. Chem. Soc. 1997, 119, 1147-1148; T. Schrader, Chem. Eur. J. 1997, 3, 1537-1541; artificial receptor molecules for a,w-diammonium ions: C. Pascard, C. Riche, M. Cesario, F. Kotzyba-Hibert, J.-M. Lehn, J. Chem. Soc. Chem. Commun. 1982, 557; A. D. Hamilton, J.-M. Lehn, J. L. Sessler, J. Am. Chem. Soc. 1996, 108, 5158.
[2] T. Schrader, M. Wehner, P. Finocchiaro, S. Failla, G. Consiglio, Org. Lett. 2000, 2, 605-608.
[3] Exploratory work on this chelate complexation proved this to be a very efficient general concept: T. Schrader, Angew. Chem. 1996, 108, 2816; T. Schrader, J. Org. Chem 1998, 63, 264-272.; T. Schrader, J. Inclusion Phenom. Macrocycl. Chem. 1999, 34, 117.
[4] G. A. Consiglio, S. Failla, P. Finocchiaro, K. I. Hardcastle, M. Visi, Supramolecular Chemistry 2000, 11, 177.
[5] R. Karaman, A. Goldblum, E. Breuer, J. Chem. Soc. Perkin Trans. 1989, 765; H. Krawczyk, Synthetic Communications 1997, 27, 3151-3161.
[6] (a) Job, P. Ann. Chim. 1928, 9, 113–203. (b) Blanda, M. T.; Horner, J. H.; Newcomb, M. J. Org. Chem. 1989, 54, 4626–4636.
[7] We found this array in numerous 1:1-complexes of bisphosphonates and short diammonium cations (unpublished results); Molecular Modeling Program: CERIUS2 from Molecular Simulations Inc., Force field: Dreiding 2.21.
[8] Strong highfield-shifts of both the imidazole and the pyridine ring protons indicate p,p-interactions, supporting the binding mode of Fig. 6.
[9] (a) Schneider, H. J.; Kramer, R.; Simova, S.; Schneider, U. J. Am. Chem. Soc. 1988, 110, 6442. (b) Wilcox, C. S. in Frontiers in Supramolecular Chemistry and Photochemistry; Schneider, H.-J., Dürr, H., Eds.; VCH: Weinheim, 1991; pp. 123–143. c) Macomber, R. S. J. Chem. Educ. 1992, 69, 375–378.
[10] T. Grawe, T. Schrader, M. Gurrath , A. Kraft, F. Osterod, Org. Lett.2000, 2, 29-32.
[11] T. W. Bell, A. B. Khasanov, M. G. B. Drew, A. Filikov, T. L. James, Angew. Chem. Int. Ed. 1999, 38, 2543-2547.
[12] S. M. Ngola, P. C. Kearney, S. Mecozzi, K. Russell, D. A. Dougherty, J. Am. Chem. Soc. 1999, 121, 1192-1201.
All comments on this poster should be sent by e-mail to (mailto:[email protected] ona.edu)
[email protected] with C0026 as the message subject of your e-mail.